The aim of our research is gaining a deeper understanding of the molecular basis and the clinical relevance of allergen-specific antibody responses. In accordance with the group's name, we employ a synergistic approach combining wet-lab experiments and in silico analyses to contribute to the progress in diagnosis and treatment of allergy.
The hallmark of immediate-type allergies is the formation of immunoglobulin E (IgE) antibodies. A long-discussed problem in allergy diagnostics is the fact that not all patients whose IgE binds to certain allergens show allergic reactions to the respective allergen source. Several factors that determine the clinical relevance of allergen-specific IgE have been discussed, including the concentrations of allergen-specific antibodies of other subclasses, such as IgG, the binding strength (avidity) of the IgE antibodies, and the number and distribution of binding sites (epitopes) recognized by IgE and IgG.
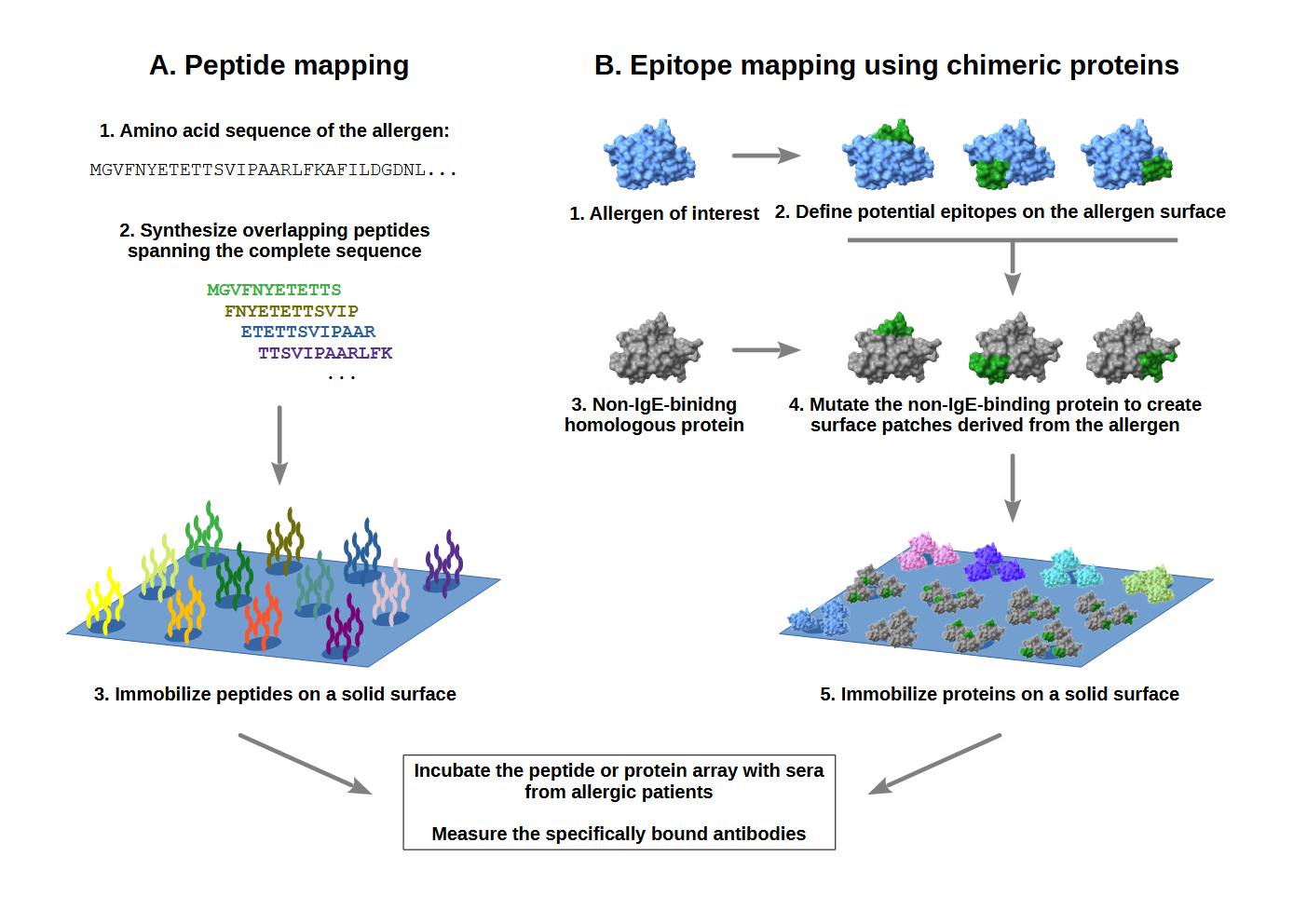
Epitopes are usually identified by testing allergic patients' sera with a panel of short, overlapping synthetic peptides whose sequences cover the complete sequence of the allergen (Figure 1, left). However, respiratory allergens get into contact with the immune system as intact, folded proteins. Therefore, most allergen-specific antibodies bind to epitopes that are present only in the folded proteins and cannot be represented by short peptides. Hence, we have developed an alternative approach based on artificial chimeric proteins that carry distinct patches derived from the surface of the allergen (Figure 1, right).
We are currently applying this approach to characterizing epitope binding patterns of allergen-specific antibodies from patients allergic to birch and grass pollen. In birch pollen allergy, we want to understand why the patients show very different patterns of allergic reactions to certain plant foods, such as apple, hazelnut, stone-fruits are carrot, although IgE antibodies of most patients bind to the major birch pollen allergen, Bet v 1, and cross-react with Bet v 1-related allergens in those plants. In grass pollen allergy, we aim at monitoring the development of epitope recognition patterns of allergen-specific IgE and IgG antibodies during the first year of allergen immunotherapy (AIT). Thereby, we hope to gain deeper insights into the immunologic mechanisms of AIT and the causes underlying the varying efficacy of the therapy.
Selected publications
- Schmalz S, Mayr V, Shosherova A, Gepp B, Ackerbauer D, Sturm G, Bohle B, Breiteneder H, Radauer C (2022): Isotype-specific binding patterns of serum antibodies to multiple conformational epitopes of Bet v 1. J Allergy Clin Immunol 149: 1786-94. [Free full text]
- Tscheppe A, Palmberger D, van Rijt L, Kalic T, Mayr V, Palladino C, Kitzmüller C, Hemmer W, Hafner C, Bublin M, van Ree R, Grabherr R, Radauer C, Breiteneder H (2020): Development of a novel Ara h 2 hypoallergen with no IgE binding or anaphylactogenic activity. J Allergy Clin Immunol 145: 229-38. [Free full text]
One of the unresolved problems in allergy research is the question of what makes a protein an allergen. In addition to studying the behavior of immune cells or animal models when exposed to allergenic and non-allergenic proteins, the availability of large allergen databases combined with sequence, structure, and protein family data now allows researchers to gain new insights through sequence and structural analysis, leading to new experimental approaches.
We are attempting to answer some of the following questions. What is the distribution of allergenic and non-allergenic proteins within allergen-containing protein families and superfamilies? Which sequence-related factors, such as similarity to human homologues, similarity to parasite or bacterial proteins, or sequence conservation within a protein family, show a correlation with allergenicity? How reliably can IgE cross-reactivity between homologous allergens be predicted based on sequence similarity data?
Selected publications
- Radauer C, Lackner P, Breiteneder H (2008): The Bet v 1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol 8: 286. [Free full text]
Most allergenic proteins can be classified into an astonishingly small number of families. Members of these protein families possess similar structures and, in many cases, similar biochemical functions. This led to the hypothesis that allergenicity depends on structural or functional characteristics of proteins. In addition, common protein family membership is a prerequisite of IgE and T cell cross-reactivity.
In order to help researchers and clinicians getting an overview of families of allergens and their allergenic members, we developed the AllFam allergen family database, a Web resource that enables the user to get protein family data for allergens compiled from the WHO/IUIS Allergen Nomenclature Database and AllergenOnline.
Selected publications
- Radauer C, Breiteneder H (2019): Allergen databases - A critical evaluation. Allergy 74: 2057-2060. [Free full text]
- Radauer C, Bublin M, Wagner S, Mari A, Breiteneder H (2008): Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J Allergy Clin Immunol 121: 847-52. [Full text]

Christian Radauer
Group leaderAndrea Martschini
PhD studentPhone: +43 1 40400 51170
Email: andrea.martschini (at) meduniwien.ac.at
Nadine Prammer
Laboratory technicianPhone: +43 1 40400 51160
Email: nadine.prammer (at) meduniwien.ac.at
Petra Natascha Hendler
Administrative assistantAlumni
- Stefanie Ottersbach (PhD Student 2018-22)
- Vanessa Mayr (Biomedical Assistant 2019-21)
- Gerlinde Hofstetter (Biomedical Assistant 2010-14)
- Eva Rauch-Guhsl (PhD Student 2010-14)
Christian Radauer: Curriculum Vitae
Christian Radauer studied Biochemistry at the University of Vienna. He performed his PhD studies in the group of Heimo Breiteneder at the former Department of General and Experimental Pathology of the University of Vienna (which is now the Department of Pathophysiology and Allergy Research of the Medical University of Vienna). Christian received his PhD degree in 2000. He continued his career in Heimo Breiteneder's lab as a post-doctoral scientist. During a short research stay in Peter Lackner's group at the Department of Molecular Biology, University of Salzburg, he gained additional experience in bioinformatics. In 2009, he received the teaching qualification (Habilitation) in molecular allergology.
Christian Radauer's research interests have focused on the field of molecular allergology. The main topics are elucidating molecular properties of allergies, factors that make certain proteins allergenic, the molecular basis of cross-reactivity, and the clinical relevance of patient specific repertoires of allergen specific immunoglobulins. Since April 2022, Christian Radauer has been scientific co-chair of the WHO/IUIS Allergen Nomenclature Sub-Committee. This function of this committee is to develop an official, unambiguous nomenclature for allergenic proteins and to make the official allergen names accessible to the scientific community in the WHO/IUIS Allergen Nomenclature Database.